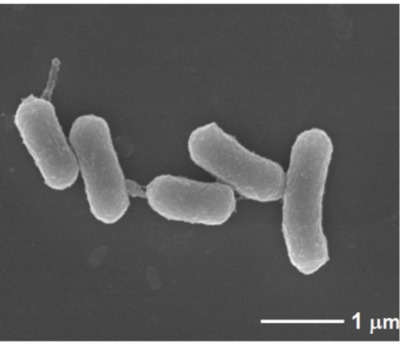
| Species: | Porphyromonas asaccharolytica |
|---|---|
| Genus: | Porphyromonas |
| Family: | Porphyromonadaceae |
| Order: | Bacteroidales |
| Class: | Bacteroidia |
| Phylum: | Bacteroidetes |
| Disease Association: |
Colorectal cancer (ES=0.326433) Colorectal cancer (ES=0.328432) Colorectal cancer (ES=0.339309) |
|---|
| Shape | Rod-shaped |
|---|---|
| Gram staining | Gram- |
| Motility | Nonmotile |
| Oxygen Requirement | Anaerobe |
| Sporulation | Nonsporulating |
| Ecosystem | Unclassified; Human |
| Ecosystem Type | Unclassified; Reproductive system |
In the linked pathways:
red=enriched, blue=depleted
ko00020 - Citrate cycle (TCA cycle)
ko00471 - D-Glutamine and D-glutamate metabolism
ko00473 - D-Alanine metabolism
ko00521 - Streptomycin biosynthesis
ko00550 - Peptidoglycan biosynthesis
ko00670 - One carbon pool by folate
ko00710 - Carbon fixation in photosynthetic organisms
ko00730 - Thiamine metabolism
ko00780 - Biotin metabolism
ko00785 - Lipoic acid metabolism
ko00970 - Aminoacyl-tRNA biosynthesis
ko03430 - Mismatch repair
ko03440 - Homologous recombination
M00005 - PRPP biosynthesis, ribose 5P => PRPP
M00007 - Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P
M00020 - Serine biosynthesis, glycerate-3P => serine
M00060 - Lipopolysaccharide biosynthesis, KDO2-lipid A
M00086 - beta-Oxidation, acyl-CoA synthesis
M00096 - C5 isoprenoid biosynthesis, non-mevalonate pathway
M00115 - NAD biosynthesis, aspartate => NAD
M00116 - Menaquinone biosynthesis, chorismate => menaquinol
M00119 - Pantothenate biosynthesis, valine/L-aspartate => pantothenate
M00122 - Cobalamin biosynthesis, cobinamide => cobalamin
M00123 - Biotin biosynthesis, pimeloyl-ACP/CoA => biotin
M00124 - Pyridoxal biosynthesis, erythrose-4P => pyridoxal-5P
M00149 - Succinate dehydrogenase, prokaryotes
M00153 - Cytochrome bd ubiquinol oxidase
M00525 - Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine
M00526 - Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine
M00565 - Trehalose biosynthesis, D-glucose 1P => trehalose
M00573 - Biotin biosynthesis, BioI pathway, long-chain-acyl-ACP => pimeloyl-ACP => biotin
M00577 - Biotin biosynthesis, BioW pathway, pimelate => pimeloyl-CoA => biotin
M00705 - Multidrug resistance, efflux pump MepA
M00793 - dTDP-L-rhamnose biosynthesis
Undetected
Undetected
Undetected
MATLAB species model file: msp_1570.mat