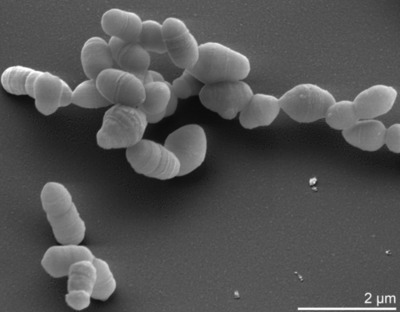
| Species: | Slackia piriformis |
|---|---|
| Genus: | Slackia |
| Family: | Eggerthellaceae |
| Order: | Eggerthellales |
| Class: | Coriobacteriia |
| Phylum: | Actinobacteria |
| Gut outflow: | 1 |
|---|
| Shape | Rod-shaped |
|---|---|
| Gram staining | Gram+ |
| Motility | Nonmotile |
| Oxygen Requirement | Anaerobe |
| Sporulation | Nonsporulating |
| Ecosystem | Human |
In the linked pathways:
red=enriched, blue=depleted
ko00290 - Valine, leucine and isoleucine biosynthesis
ko00300 - Lysine biosynthesis
ko00471 - D-Glutamine and D-glutamate metabolism
ko00473 - D-Alanine metabolism
ko00550 - Peptidoglycan biosynthesis
ko00670 - One carbon pool by folate
ko00730 - Thiamine metabolism
ko00750 - Vitamin B6 metabolism
ko00770 - Pantothenate and CoA biosynthesis
ko00970 - Aminoacyl-tRNA biosynthesis
M00005 - PRPP biosynthesis, ribose 5P => PRPP
M00017 - Methionine biosynthesis, apartate => homoserine => methionine
M00019 - Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine
M00020 - Serine biosynthesis, glycerate-3P => serine
M00050 - Guanine ribonucleotide biosynthesis IMP => GDP,GTP
M00140 - C1-unit interconversion, prokaryotes
M00153 - Cytochrome bd ubiquinol oxidase
M00157 - F-type ATPase, prokaryotes and chloroplasts
M00365 - C10-C20 isoprenoid biosynthesis, archaea
M00432 - Leucine biosynthesis, 2-oxoisovalerate => 2-oxoisocaproate
M00525 - Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine
M00535 - Isoleucine biosynthesis, pyruvate => 2-oxobutanoate
M00705 - Multidrug resistance, efflux pump MepA
M00844 - Arginine biosynthesis, ornithine => arginine
Undetected
Undetected
Undetected
Undetected
MATLAB species model file: msp_1634.mat